Function performing a test that evaluates the null hypothesis of block-independence against the alternative of block-dependence (presence of non-zero elements in the off-diagonal block) in the precision matrix using high-dimensional data. The mentioned test is a permutation-based test (see details).
GGMblockTest(
Y,
id,
nPerm = 1000,
lambda,
target = default.target(covML(Y)),
type = "Alt",
lowCiThres = 0.1,
ncpus = 1,
verbose = TRUE
)Arguments
- Y
Data
matrix. Variables assumed to be represented by columns.- id
A
numericvector acting as an indicator variable for two blocks of the precision matrix. The blocks should be coded as0and1.- nPerm
A
numericorintegerdetermining the number of permutations.- lambda
A
numericrepresenting the penalty parameter employed in the permutation test.- target
A target
matrix(in precision terms) for Type I ridge estimators.- type
A
characterindicating the type of ridge estimator to be used. Must be one of: "Alt", "ArchI", "ArchII".- lowCiThres
A
numerictaking a value between 0 and 1. Determines speed of efficient p-value calculation.- ncpus
A
numericorintegerindicating the desired number of cpus to be used.- verbose
A
logicalindicating if information on progress and output should be printed on screen.
Value
An object of class list:
- statistic
A
numericrepresenting the observed test statistic (i.e., likelihood ratio).- pvalue
A
numericgiving the p-value for the block-independence test.- nulldist
A
numericvector representing the permutation null distribution for the test statistic.- nperm
A
numericindicating the number of permutations used for p-value calculation.- remark
A
"character"that states whether the permutation algorithm was terminated prematurely or not.
Details
The function performs a permutation test for the null hypothesis of block-independence against the alternative of block-dependence (presence of non-zero elements in the off-diagonal block) in the precision matrix using high-dimensional data. In the low-dimensional setting the common test statistic under multivariate normality (cf. Anderson, 2003) is: $$ \log( \| \hat{\mathbf{\Sigma}}_a \| ) + \log( \| \hat{\mathbf{\Sigma}}_b \| ) - \log( \| \hat{\mathbf{\Sigma}} \| ), $$ where the \(\hat{\mathbf{\Sigma}}_a\), \(\hat{\mathbf{\Sigma}}_b\), \(\hat{\mathbf{\Sigma}}\) are the estimates of the covariance matrix in the sub- and whole group(s), respectively.
To accommodate the high-dimensionality the parameters of interest are
estimated in a penalized manner (ridge-type penalization, see
ridgeP). Penalization involves a degree of freedom (the
penalty parameter: lambda) which needs to be fixed before testing. To
decide on the penalty parameter for testing we refer to the
GGMblockNullPenalty function. With an informed choice on the
penalty parameter at hand, the null hypothesis is evaluated by permutation.
Hereto the samples are permutated within block. The resulting permuted data
set represents the null hypothesis. Many permuted data sets are generated.
For each permutation the test statistic is calculated. The observed test
statistic is compared to the null distribution from the permutations.
The function implements an efficient permutation resampling algorithm (see
van Wieringen et al., 2008, for details.): If the probability of a p-value
being below lowCiThres is smaller than 0.001 (read: the test is
unlikely to become significant), the permutation analysis is terminated and
a p-value of unity (1) is reported.
When verbose = TRUE also graphical output is generated: A histogram
of the null-distribution. Note that, when ncpus is larger than 1,
functionalities from
snowfall are imported.
References
Anderson, T.W. (2003). An Introduction to Multivariate Statistical Analysis, 3rd Edition. John Wiley.
van Wieringen, W.N., van de Wiel, M.A., and van der Vaart, A.W. (2008). A Test for Partial Differential Expression. Journal of the American Statistical Association 103: 1039-1049.
See also
Examples
## Obtain some (high-dimensional) data
p = 15
n = 10
set.seed(333)
X = matrix(rnorm(n*p), nrow = n, ncol = p)
colnames(X)[1:15] = letters[1:15]
id <- c(rep(0, 10), rep(1, 5))
## Generate null distribution of the penalty parameter
lambda0dist <- GGMblockNullPenalty(X, id, 5, 0.001, 10)
## Location of null distribution
lambdaNull <- median(lambda0dist)
## Perform test
testRes <- GGMblockTest(X, id, nPerm = 100, lambdaNull)
#> 0 of 100 permutations done, and counting...
#> 25 of 100 permutations done
#> 25 of 100 permutations done, and counting...
#> 50 of 100 permutations done
#> 50 of 100 permutations done, and counting...
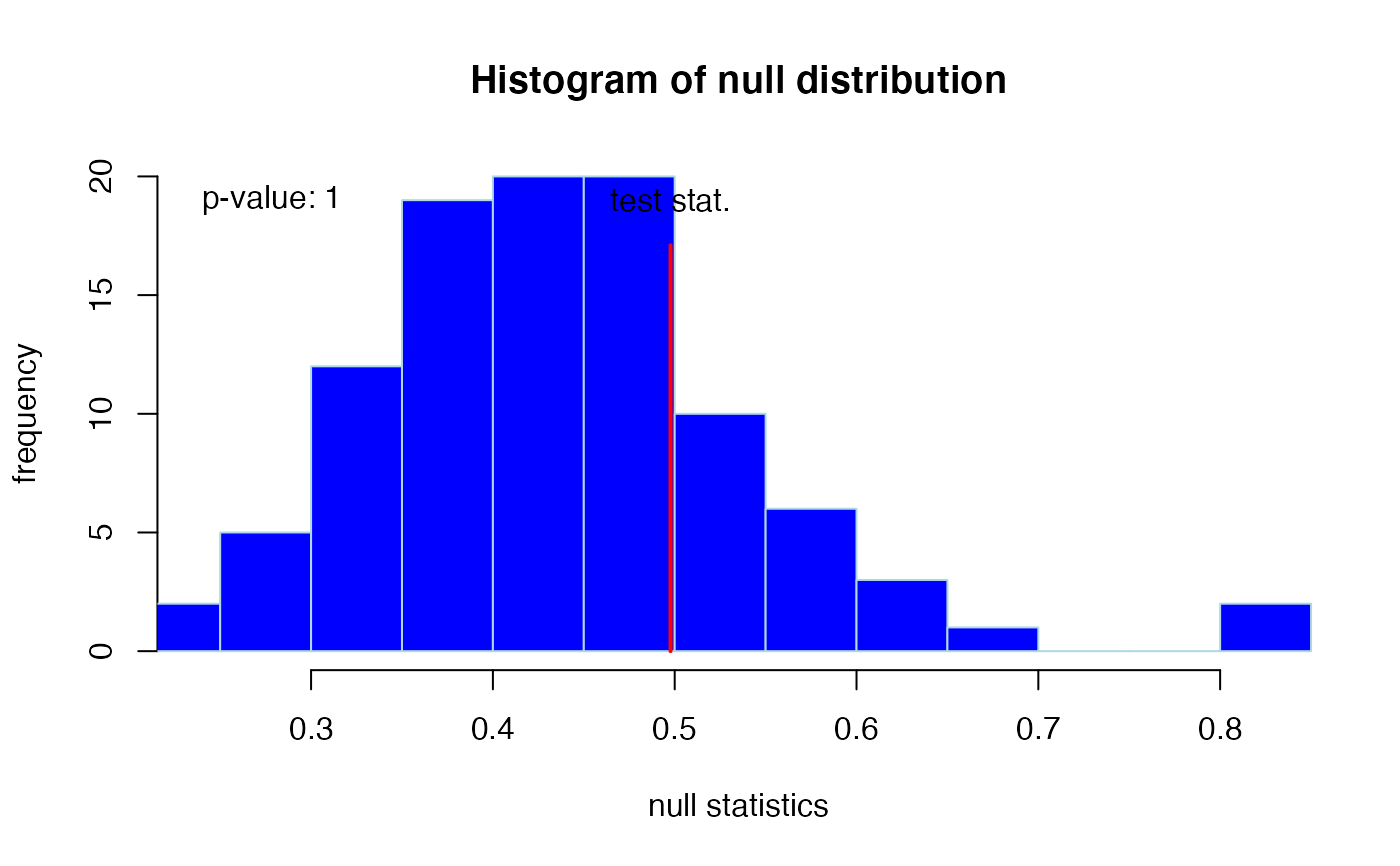 #>
#> Likelihood ratio test for block independence
#> ----------------------------------------
#> -> number of permutations : 100
#> -> test statistic : 0.498
#> -> p-value : 1
#> -> remark : resampling terminated prematurely due to unlikely significance
#> ----------------------------------------
#>
#>
#> Likelihood ratio test for block independence
#> ----------------------------------------
#> -> number of permutations : 100
#> -> test statistic : 0.498
#> -> p-value : 1
#> -> remark : resampling terminated prematurely due to unlikely significance
#> ----------------------------------------
#>